RESEARCH TEAMS
HUMAN SYSTEMS IMMUNOLOGY & INFLAMMATORY NETWORKS TEAM
The interdisciplinary team works exclusively in the human system to better understand its function in physiology and disease. Using translational approaches, in overlapping studies ,the team provides a state-of-the-art and internationally recognized expertise in human clinical and basic research.
PROJECTS
Summary of the team main achievements
The strong interface between systems biology and immunology is now internationally recognized through high level publications (in Cell, Nat Cell Biol, Nat Immunol), and coordination of high-impact consortia. The team takes pride of:
- 128 original peer reviewed publications, ¬13 000 citations
Publications across different disease: allergy, auto-immunity, infection, cancer, COVID-19 - Creation of the DrugSynergy platform: a human pre-clinical platform for identifying rational drug combinations using high resolution analysis of cell behavior
- Generation and management of unique patient-derived datasets:
- T-MEGA (Tumor microenvironment global analysis): a resource dataset derived from >1000 human primary tumor samples, combining cellular and molecular multi-layer information
- MAARS (Microbes in Allergy and Autoimmunity Related to the Skin): a large-scale dataset of combined microbiome and transcriptome from human primary skin samples (>600 samples)
- Industrial partnerships in immuno-oncology: Roche, Sanofi, Elsalys, Servier, Epigene Labs
- Major european grants:
- ERC consolidator (2011) and Proof-Of-Concept (POC) (2015) grants
- FP7 MAARS (2011-2016): coordinator of bioinformatics and systems biology
- H2020 ImmunAID (2018-2024): project coordinator
- IMI IMMUcan (2019-2024): Bioinformatics and data integration workpackage coordinator
.
Immunobiology of dendritic cells and T cells
- Immunobiology of dendritic cells and T cells
This topic has been a focus of the team’s research since its creation in 2004. We have the expertise and know-how about purifying, culturing, stimulating, and analyzing rare DC subsets, and studying their interactions with T cells. In parallel, we integrate the most cutting-edge technologies such as 35-color flow cytometry, single-cell RNAseq, and other multiplex technologies, together with complex data analysis methods. Our laboratory continues working on human DC and T cell biology at the highest international standards, to address important immunological and systems biology questions. We have made seminal contributions to DC diversity and function in human disease, such as DC adaptation to the breast cancer microenvironment (Michea et al, Nat Immunol, 2018), and the recent single-cell resolution characterization of DC in COVID-19 (Saichi et al, Nat Cell Biol, 2021).
From immunology to systems biology
From immunology to systems biology
Complexity is present everywhere in living systems, structured according to general rules, some of them applicable to all hierarchical scales of living systems organization, some of them specific to a certain scale (cell, tissue, organ, organisms). To understand this complexity, we are asking systems biology questions, such as cell state transition, multiple signal integration, adaptation to microenvironmental changes, and cell-cell communication, using immune cells as a model. This led to international leadership in studying human immune cell communication through a combination of experimental and computational approaches. We have developed ICELLNET, a computational framework to infer cell-cell communication from single-cell or bulk transcriptomic data (Noel et Massenet-Regad et al, Nat Commun 2021).
Using a systems approach inspired by linguistics, we were the first to establish a multivariate mathematical model of cell-cell communication, using dendritic cells (DC) and T cells as a model (Grandclaudon et al, Cell 2019). At the next level, we aim at expanding the systems analysis of DC-T cell communication and providing the first basis for cellular linguistics.
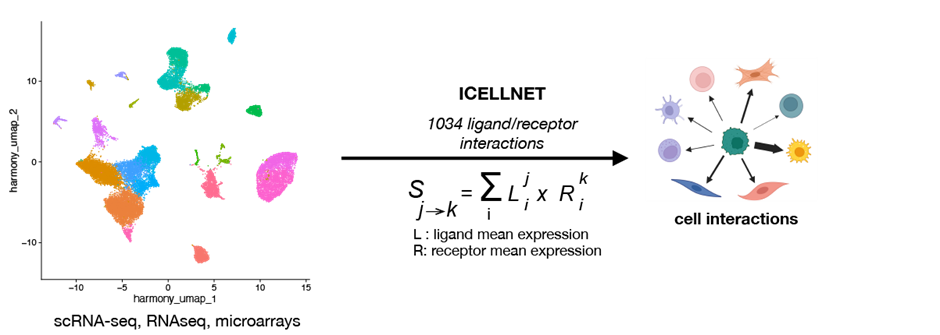
From systems approach to cell networks in cancer
-
- From systems approach to cell networks in cancer
The complexity of cancer-associated inflammation relies on diverse cell compartments interacting via soluble and surface molecules. In a systems approach, these two components could be simultaneously deciphered by computational and experimental methods, and the results then combined to elucidate cell network structure and topology.
Our systems approach initially deciphers the transcriptional diversity of DC across cancer. Next, by using high-throughput methodologies we explore how dysregulated pathways in stromal, dendritic and T cells relate to their physiopathology. Cell communication is addressed by investigating cytokine signatures. Finally, our original data will be integrated with public datasets by using computational methods resulting in the reconstruction of cell communication networks that may influence tumour growth and clinical outcome. To that aim, we have recently contributed to building a unique database of scRNA-seq cancer studies (Camps et al, Can Res 2022 ). Overall, we aim to elucidate the organization of inflammatory networks in distinct tumour types at the depth of precision medicine, offering the potential for novel immunotherapeutic developments.
The complexity of the tumour microenvironment - From systems approach to cell networks in cancer
Stromal cells in health and rare disease
-
- Stromal cells in health and rare disease
Fibroblastic reticular cells (FRCs) actively support secondary lymphoid organ (SLO) architecture. By compartmentalizing immune cells in specialized niches, and tightly interacting with them, FRC regulate innate and adaptive immunity. We comprehensively profile the phenotypic and functional diversity of FRCs in human SLOs. In parallel, we are comparing the lymphoid stromal cell network in healthy donors to the one in patients suffering from a group of rare disorders named Castleman disease. This ongoing collaboration with the National Reference Center for Castleman Disease could result in new diagnostic tools and innovative therapeutic targets.
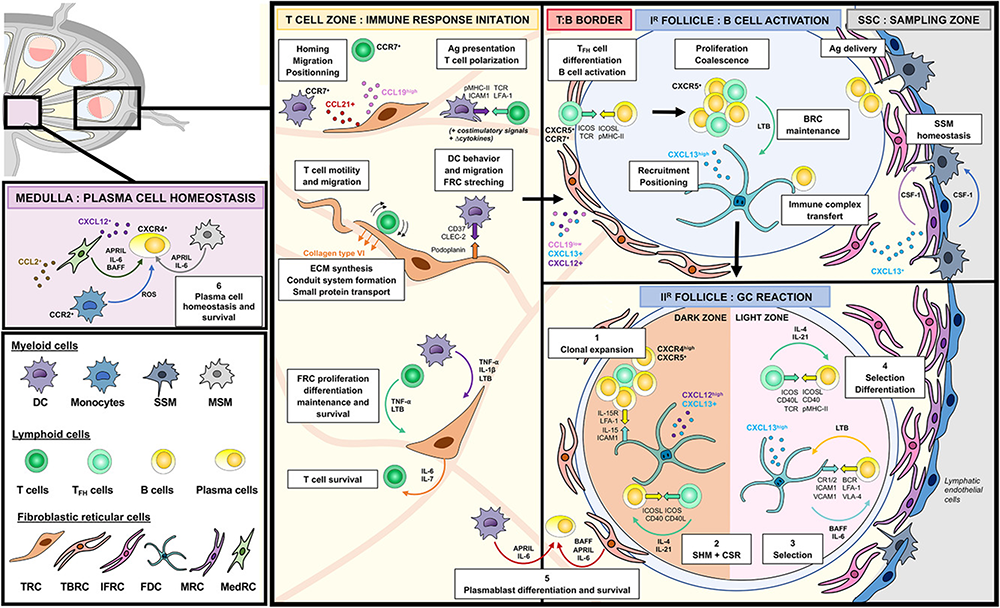
Compartmentalized multicellular crosstalk in lymph nodes coordinates the generation of potent cellular and humoral immune responses Eur J Immunol, Volume: 51, Issue: 12, Pages: 3146-3160, First published: 04 October 2021, DOI: (10.1002/eji.202048977) - Stromal cells in health and rare disease
ImmunAID
 The European H2020 ImmunAID project is seeking to deliver a method for rapid and accurate diagnosis across the spectrum of SAID (systemic autoinflammatory diseases), to improve clinical management of SAID patients. The ImmunAID project brings together 25 institutions and 35 clinical centers from 12 European countries, under the coordination of INSERM.
The European H2020 ImmunAID project is seeking to deliver a method for rapid and accurate diagnosis across the spectrum of SAID (systemic autoinflammatory diseases), to improve clinical management of SAID patients. The ImmunAID project brings together 25 institutions and 35 clinical centers from 12 European countries, under the coordination of INSERM.
A large patient cohort is being constituted with the aim to collect clinical data and biological samples from SAID patients and healthy donors. Next, the partners perform unbiased multiomics approaches (genomic, transcriptomics, proteomics and microbiomics) as well as hypothesis-driven assays exploring the inflammasome, inflammation resolution and immune networks. The clinical data and the biological data analysis will go through big data analyses via a centralized data management strategy.
The main objectives of ImmunAID are:
- To deliver a new, comprehensive and pathogenesis-driven classification of SAID
- To open the way to a rapid and accurate diagnosis across all the spectrum of SAID
- To identify and validate novel Omics- and pathway-based diagnostic biomarkers: genetic mutations (or combinations of genetic mutations), translational dysregulation, proteomics disequilibrium or microbiome alteration associated with SAID
- To describe the potential dysregulation of inflammasome functions, to assess the role of cytokine network disequilibrium in the expression of SAIDs, and to explore inflammation resolution processes.
For full information on the ImmunAID project please visit ImmunAID.eu or send a message to contact@immunaid.eu
This project received funding from the EU’s Horizon 2020 research and innovation programme under grant agreement No. 779295.
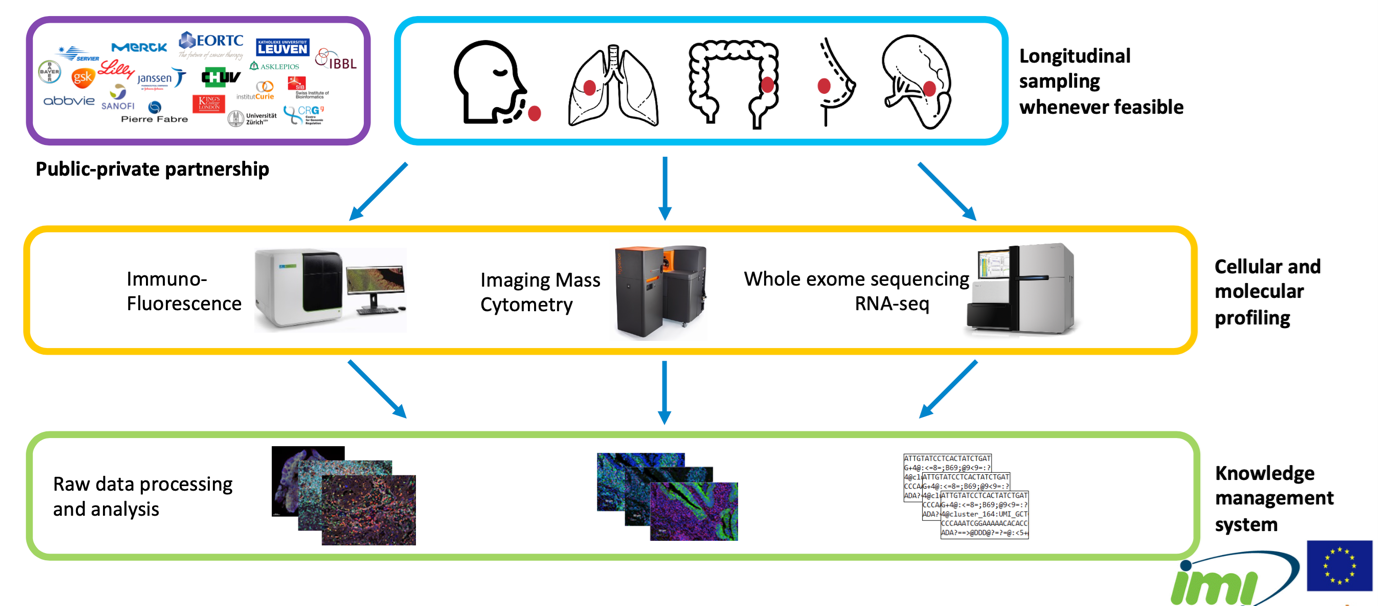
IMMUCAN
IMMUCAN
 The complexity and heterogeneity of the tumour microenvironment remains a major challenge for a better understanding of cancer development, progression and treatment. The IMMUcan consortium aims to address this need.
The complexity and heterogeneity of the tumour microenvironment remains a major challenge for a better understanding of cancer development, progression and treatment. The IMMUcan consortium aims to address this need.
The mission of IMMUcan is to generate broad molecular and cellular profiling data of the tumour microenvironment from up to 3,000 patients derived from five indications: non-small cell lung cancer, head and neck squamous cell carcinoma, renal cell carcinoma, colorectal cancer and breast cancer. All data will be integrated with clinical information to provide a better understanding of the TME and identify patients that might respond to treatment, as well as resistance mechanisms.
The IMMUcan project has received funding from the Innovative Medicines Initiative 2 Joint Undertaking under grant agreement No 821558..
Principal Investigators
- Pr. Vassili SOUMELIS, MD, PhD, Professor, Immunology (on leave)
- Dr. Evanguelos Xylinas, MD, PhD, FEBU, Maître de Conférences des Universités, Urology service, Bichat-Claude Bernard Hospital

Latest publications

Latest news
Job openings
Jul 1, 2021


